AI-Driven Peptide Biomarker Discovery for Tuberculosis
Tuberculosis (TB) remains a major global health concern, claiming millions of lives each year. The traditional methods of diagnosing TB, while effective, have limitations in terms of speed and accuracy. Enter microarray technology—a powerful tool that allows for the concurrent measurement of thousands of gene expressions, providing a comprehensive genomic profile of diseases.
In this digital era, harnessing the power of Artificial Intelligence alongside microarray data enables the offering of innovative solutions for TB diagnosis. Our platform is designed to streamline the analysis process, transform complex data into actionable insights, and harness peptide microarray data and computational intelligence to distinguish TB from non-TB immune responses with precision.
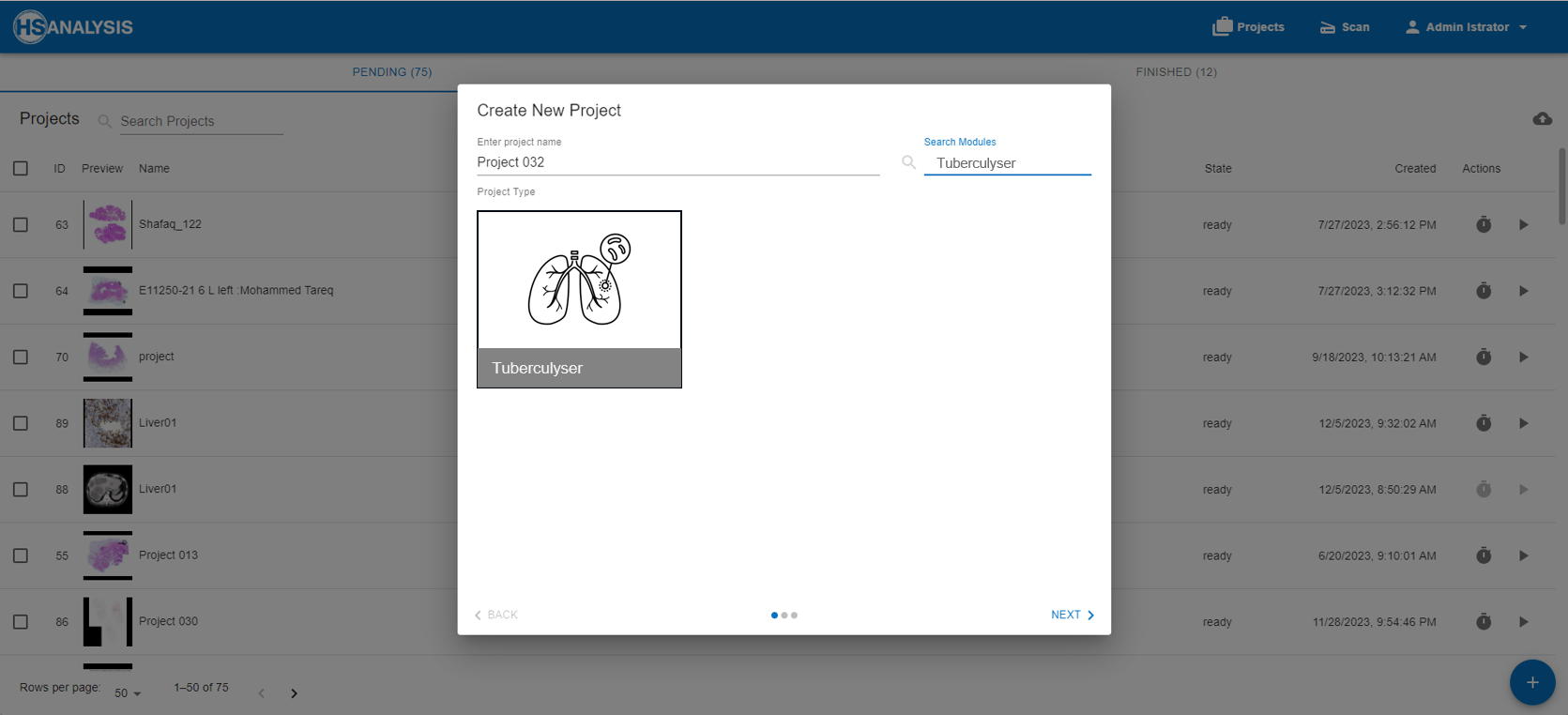
Working with HSA KIT
HSA KIT provides a solution for developing and training custom deep learning or machine learning models. With hundreds of scalable modules available, this software is built to meet long-term requirements and create flexible solutions to adapt evolving needs of our clients.
All the essential tools, from data preprocessing to model building and evaluation, can be easily accessed by a user without prior technical knowledge. The user-friendly interface and comprehensive documentation make it accessible to even beginners, allowing anyone to learn and leverage this powerful tool quickly.
About the Project: Tuberculyzer
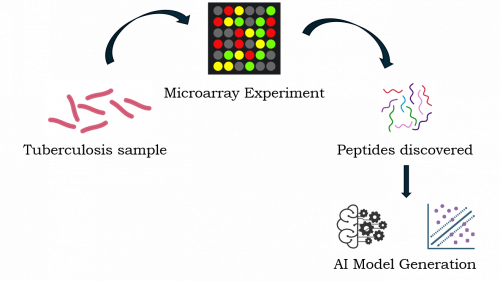
Tuberculyzer in HSA KIT leverages peptide microarray technology and machine learning to identify antibody-based signatures that distinguish TB-positive from non-TB samples. This integrative approach combines bioinformatics, statistical peptide profiling, and artificial intelligence to uncover new diagnostic biomarkers.
Scientific Background
| 🧪 Peptide Microarrays | Microarray experiments measure the expression levels of thousands of genes or proteins at once. In the case of peptide biomarker discovery, this involves a peptide microarray, where a patient’s serum is screened against a library of potential peptide antigens |
| 🤖 Machine Learning Integration | The AI model can identify which peptides show the most significant differences in binding patterns between patients with and without TB. This helps narrow down the list of potential biomarkers from thousands of candidates to a manageable few. |
| 🔍 Biomarker Discovery | Algorithms are trained on the data to classify patients as TB (active or latent) or non-TB (healthy). By doing this, the models learn to identify the “peptide signature” of each disease state |
| 🌍 Goal | Enable faster, minimally invasive, and data-driven TB diagnosis using blood-based peptide signatures |
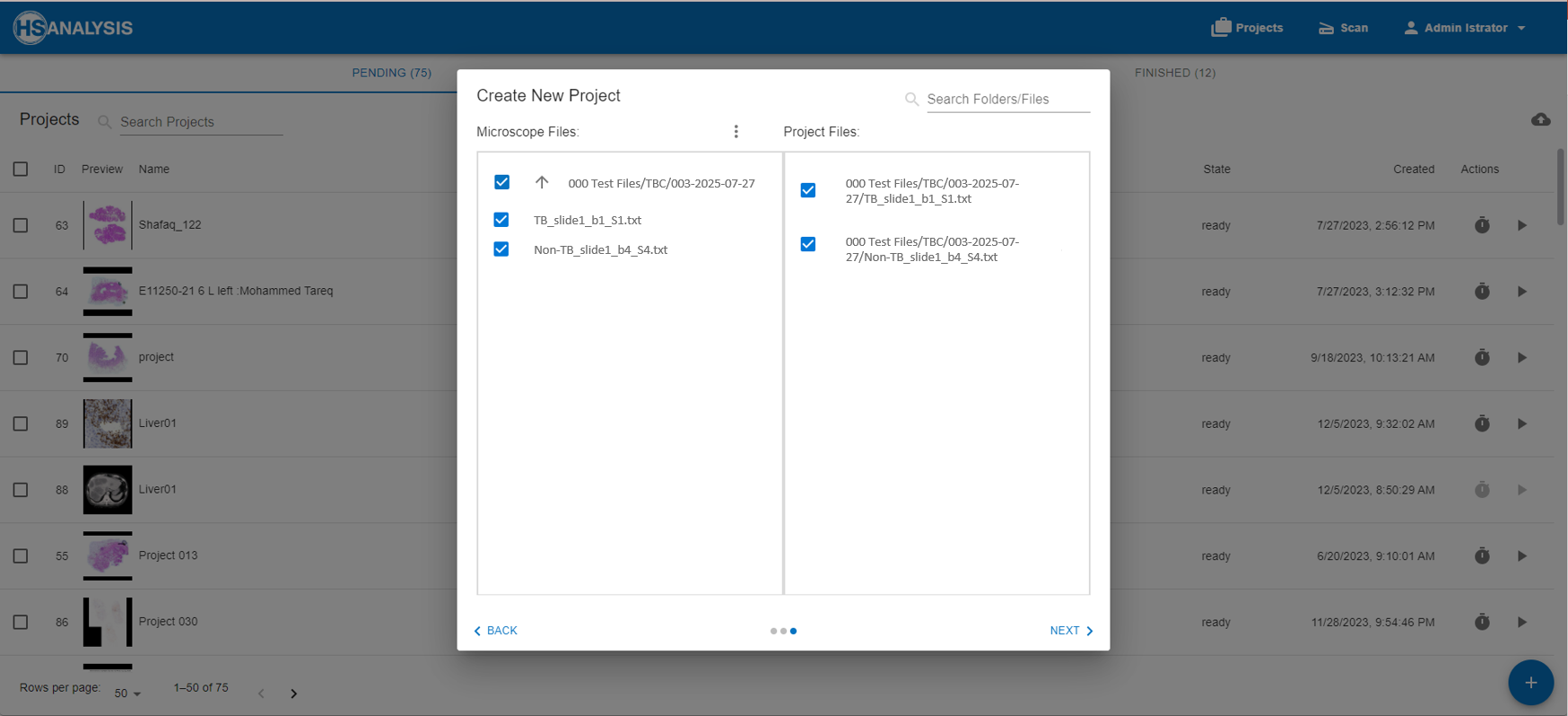
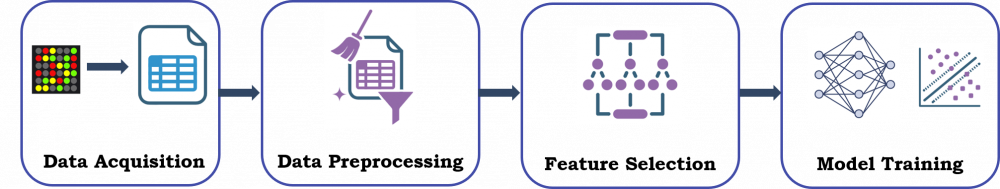
Our Workflow: From Raw Data to Predictive Modelling
- The raw data obtained from microarray experiments was acquired for both TB and non-TB samples
- Data Preprocessing included background correction, normalization, and quality filtering to remove noisy or empty spots
- We used Random Forest feature selection to identify parameters which were significantly different between the two datasets
- A Support Vector Machine classifier was trained on only the peptides with significantly different intensities between the two datasets
- Model tested on an independent validation set and evaluated using AUC, sensitivity, and specificity
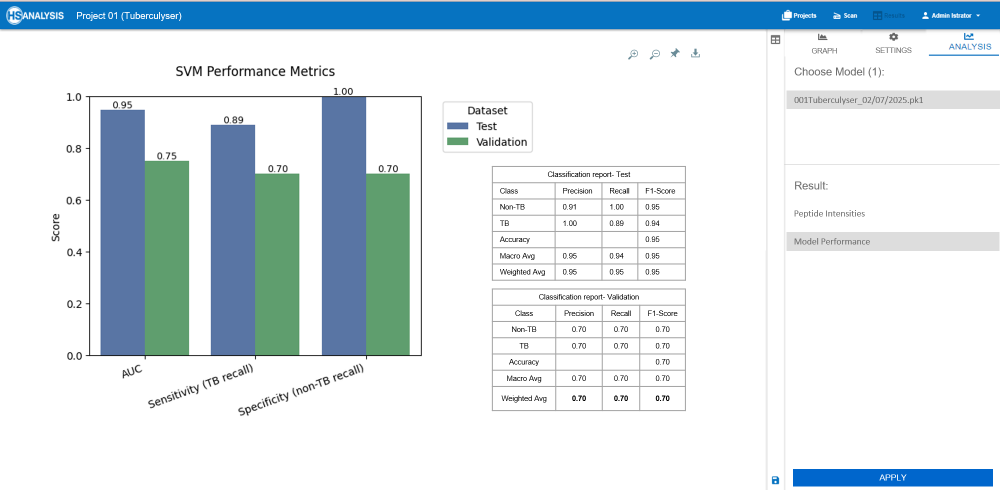
Results from Tuberculyser
-
Strong overall performance:
The model shows excellent ability to distinguish between TB and non-TB samples, with a Test AUC of 0.947 and Validation AUC of 0.75 -
Clinically meaningful sensitivity:
Achieving 70% sensitivity on validation data is a solid result for biological samples, where variability between patients and sample quality often limits perfect detection -
Reliable for screening workflows:
The model’s balance of sensitivity and specificity makes it suitable as a pre-screening or triage tool, identifying likely TB cases for confirmatory testing
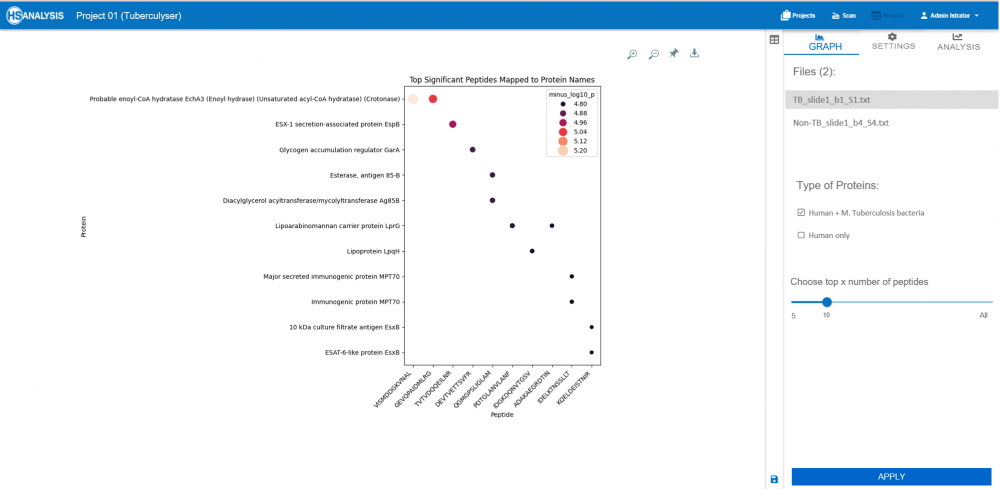
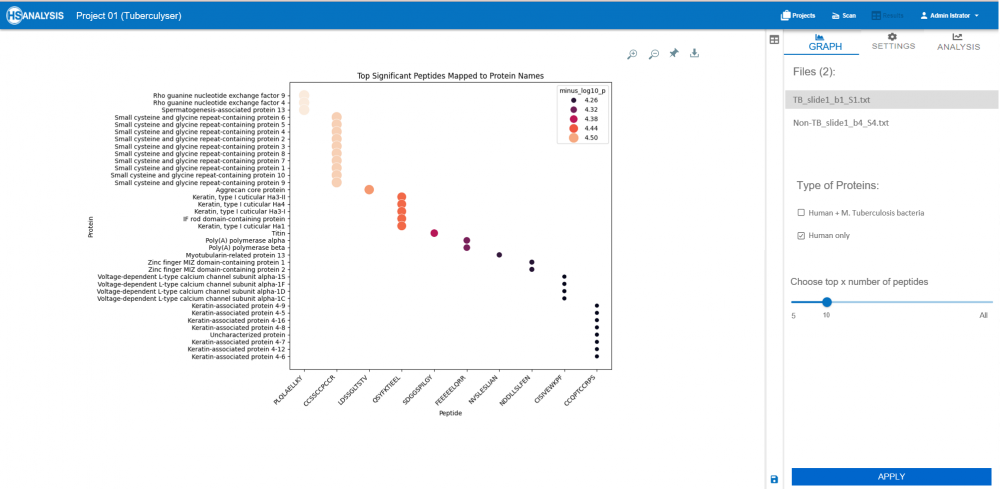
During TB infection, the host’s immune and inflammatory systems respond to the invading bacteria. To understand these biological changes, the significantly different peptides (those that changed notably between TB and non-TB samples) were traced back to the proteins they originated from
These proteins belonged either to:
-
-
Mycobacterium tuberculosis (the pathogen) — revealing bacterial components active during infection
-
Humans (the host) — highlighting immune or inflammatory processes altered in response to TB
By mapping peptides to their respective proteins, we could link molecular changes to biological functions, helping us see how TB infection affects both the bacteria’s activity and the human immune response
-
Biological Interpretation
The significant peptides mapped to well-established TB antigens such as ESAT-6, Ag85B, and EspB, known for eliciting robust immune responses in active TB cases. This validates the reliability of the peptide microarray approach in detecting infection-specific antibody profiles.
Simultaneously, the host-reactive peptides (e.g., keratins, RhoGEFs, calcium channels) suggest broader immunopathological effects and tissue responses during infection. Together, these findings demonstrate that peptide-level immune signatures can serve as diagnostic fingerprints for TB.
Applications
The Tuberculyzer module in HSA KIT provides a scalable, data-driven diagnostic framework that bridges experimental biology with computational precision.
Applications:
🏥 Clinical Diagnostics: Rapid identification of TB cases from serum antibody profiles.
🔬 Research: Discovery of novel peptide biomarkers for TB immunology.
🌐 Public Health Surveillance: Monitoring immune trends in high-burden populations.
💊 Drug and Vaccine Development: Evaluating immune responses to vaccine candidates or therapeutics.